![]()
|
|
| Beitar 400, 83 - 85 (1999) © Tel Hay Publishers Ltd. |
|
SHARON G. WOLF*, DAPHNA FRENKIEL*, TALMON ARAD +, STEVEN E. FINKEL!, ROBERTO KOLTER! & ABRAHAM MINSKY*
The crystalline state is considered to be incompatible with life. However, in living systems exposed to severe environmental assaults, the sequestration of vital macromolecules in intracellular crystalline assemblies may provide an efficient means for protection. Here we report a generic defence strategy found in Escherichia coli, involving co-crystallization of its DNA with the stress-induced protein Dps1,2. We show that when purified Dps and DNA interact, extremely stable crystals form almost instantaneously, within which DNA is sequestered and effectively protected against varied assaults. Crystalline structures with similar lattice spacings are formed in E. coli in which Dps is slightly over expressed, as well as in starved wild-type bacteria. Hence, DNA-Dps co-crystallization is proposed to represent a binding mode that provides wide-range protection of DNA by sequestration. The rapid induction and large-scale production of Dps in response to stress, as well as the presence of Dps homologues in many distantly related bacteria, indicate that DNA protection by biocrystallization may be crucial and widespread in prokaryotes.
Under conditions of either oxidative or nutritional stress, E. coli produces high levels of the nonspecific DNA-binding protein Dps2,3,4, which effectively protects DNA against oxidative agents both in vitro and in vivo4. Dps spontaneously forms spherical dodecamers that are homologous to assemblies formed by ferritins5. It has consequently been proposed that the protective activity of Dps stems from its putative ability to sequester iron ions5. While effective in attenuating formation of free radicals by the Fenton reaction, a defence strategy based solely on ion sequestration cannot provide the all-encompassing DNA protection that is required--and observed--during prolonged starvation1,6. Indeed, Dps protects DNA against various nucleases2 and oxidative agents4, thus indicating a general Dps-dependent defence mechanism.
Details of this mechanism were derived from electron microscopy studies of Dps and Dps-DNA complexes (Fig. 1). In the absence of DNA, Dps forms spherical dodecameric particles of  90 Å in diameter2, which appear ring-like in projection. Overnight incubation of Dps results in two-dimensional crystals; projection images of these crystals reveal hexagonal packing with a spacing of
90 Å in diameter2, which appear ring-like in projection. Overnight incubation of Dps results in two-dimensional crystals; projection images of these crystals reveal hexagonal packing with a spacing of 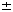 1 Å
1 Å

Figure 1 Electron microscopy of Dps and Dps-DNA complexes.
Full legend
High resolution image and legend (548k)
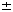 2 Å
2 ÅComplexes between DNA and nonspecific DNA-binding proteins such as RecA8, H-NS9, the sporal SASP10, sperm protamine11 and histone H1 (ref. 12) contain only protein-coated DNA molecules or amorphous aggregates. Thus, the fast DNA-Dps co-crystallization represents an unprecedented binding and protection mode. But is this mode physiologically relevant?
Electron microscopy studies were conducted on E. coli that overexpress Dps4. Over-production of Dps to levels that are fourfold higher than those accumulated in starved wild-type bacteria results in the appearance of intracellular crystalline structures (Fig. 2a-d). The flat in vitro crystals adopt a favoured orientation on the electron microscope grid and hence exhibit only intra-layer spacings. In contrast, information on both intra- and inter-layer spacings is provided by the in situ crystalline structures, as they are embedded within randomly sliced sections. Indeed, some of these crystals have a layered morphology, whereas others display square lattices that result from views normal to the layer planes. Although they are smaller and less frequent than the crystals formed following Dps over-production, crystalline assemblies are also detected in starved wild-type E. coli (Fig. 2e), thus indicating that ordered assemblies are not merely a consequence of Dps over-expression. The similarity between the in vitro and in vivo DNA-Dps crystals indicates that the ordered intracellular assemblies are Dps-DNA complexes. Several lines of evidence support this idea. First, dps knockout bacteria do not show crystalline assemblies upon starvation (Fig. 2f). Thus, the presence and intracellular amounts of Dps correlate with the appearance, size and frequency of the ordered in vivo assemblies. Second, when DNA is isolated from bacteria over-producing Dps, Dps is found to be the predominant protein bound to the DNA. Finally, in actively growing bacteria, chromatin is detected as amorphous spaces13. The absence of such regions in bacteria that contain crystalline assemblies implies a co-localization of DNA and the crystals.
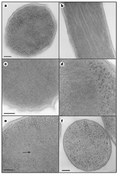 |
Figure 2 In situ DNA-Dps assemblies.
Full legend High resolution image and legend (452k) |
Both wild-type and over-producer strains form ordered structures only when starved, when Dps is one of the few proteins still being expressed2. This observation points to a causal link between starvation and the crystallization process. Because after prolonged starvation the cellular content of most proteins is reduced14, any interference that might be imposed by competing DNA-binding proteins upon the ordered DNA-Dps assembly is substantially attenuated.
The stress-related intracellular assembly and the rapid DNA-Dps co-crystallization observed in vitro imply a drive for ordered packing that is rarely encountered in living systems or revealed by biopolymers. One intriguing example is the crystalline organization of ferritin in the bacterium Helicobacter pylori15. This organization has been proposed to reduce the risk of iron poisoning by sequestering intracellular iron. The observation that both ferritin and Dps form intracellular crystalline assemblies is provocative in light of their structural homology5,16,17 and their abundance in diverse prokaryotes17,18,19,20,21. These findings may indicate an evolutionary pressure to promote biocrystallization processes whose protective activity is exerted through a fast sequestration of damaging agents and their targets.
Methods
Dps purification. We purified Dps from a Dps over-producer E. coli strain (SF101) that harbours plasmids in which the dps gene was cloned under the control of the arabinose-inducible PBAD promoter4. Cells were grown at 37 °C in LB medium to an absorbance at 560 nm of  1 and induced with 0.3% arabinose. Purification was performed as described2, with several modifications. Specifically, we replaced the Sephadex G-100 and Sepharose 6B columns with a DEAE-cellulose ion-exchange column. The final purification step remained DNA-cellulose affinity chromatography.
1 and induced with 0.3% arabinose. Purification was performed as described2, with several modifications. Specifically, we replaced the Sephadex G-100 and Sepharose 6B columns with a DEAE-cellulose ion-exchange column. The final purification step remained DNA-cellulose affinity chromatography.
DIC and fluorescence microscopy. Dps-DNA complexes (78 µg ml-1 Dps, linearized pBluescript DNA in 10 mM NaCl, 10 mM Tris) were prepared at a Dps/DNA w/w ratio of 1/5. DNA was labelled with YOYO fluorophore before complex formation at a 1/1 molar ratio of dye to base pair (bp). Images in both DIC and fluorescence modes were taken on an Axiovert 35 mm Zeiss microscope with an enhanced Gatan CCD camera.
In vitro electron microscopy. We incubated samples containing Dps and DNA (closed circular supercoiled or linearized pBluescript, 2,958 bp) at concentrations indicated in the caption to Fig. 1 at room temperature in 20 mM NaCl, 10 mM Tris (pH 7.2) on carbon-coated grids. Specimens were stained with 1% uranyl acetate and probed on a Philips CM12 electron microscope with a Tietz CCD, operating at 100 kV.
Thin-section electron microscopy. Wild-type ZK126 E. coli cells2 were grown in LB medium and starved for 48 h. E. coli harbouring the pBAD-dps plasmid that carries an arabinose-inducible dps gene4 were treated with 0.02% arabinose (resulting in Dps accumulation to a level that is 3- to 4-fold higher than that obtained in starved wild-type bacteria). Following induction, we incubated the cells in LB medium at 37 °C for 48 h. Bacteria were fixed by ultra-fast freezing in liquid ethane and subsequently cryosubstituted as described13. In control experiments, chemical fixation was performed according to the RK-U procedure22. Samples were embedded in epon; thin sections were stained with 1% uranyl acetate and examined on a Philips CM12 electron microscope.
Received 15 March;![]() accepted 5 May 1999.
accepted 5 May 1999.
References
Acknowledgements. This work was supported by a grant from the Consortium of the German Chemical Association. We thank S. Levin-Zidman and E. Shimoni for their help in the electron microscopy studies, and H. Salman for his assistance in the DIC and fluorescence experiments.
Correspondence and requests for materials should be addressed to A.M. (e-mail: cominsky@weizmann.weizmann.ac.il).